I’ve added a new PubMed search feature to Elegans, the visual worm brain explorer. The idea here is to show the network of C. Elegans neurons that get mentioned in more than n papers on PubMed, in the context of a given search query. So, for example, if one is interested in the worm’s chemotaxis behaviour, one would type in ‘chemotaxis’ and choose the citation threshold n. Initiating the search will then return the neurons that get mentioned in at least n papers along with the word ‘chemotaxis’. The search is in fact performed once for each neuron …
Articles with the python tag
Analyzing tf-idf results in scikit-learn
In a previous post I have shown how to create text-processing pipelines for machine learning in python using scikit-learn. The core of such pipelines in many cases is the vectorization of text using the tf-idf transformation. In this post I will show some ways of analysing and making sense of the result of a tf-idf. As an example I will use the same kaggle dataset, namely webpages provided and classified by StumbleUpon as either ephemeral (content that is short-lived) or evergreen (content that can be recommended long after its initial discovery).
Tf-idf
As explained in the previous post, the tf-idf …
Pipelines for text classification in scikit-learn
Scikit-learn’s pipelines provide a useful layer of abstraction for building complex estimators or classification models. Its purpose is to aggregate a number of data transformation steps, and a model operating on the result of these transformations, into a single object that can then be used in place of a simple estimator. This allows for the one-off definition of complex pipelines that can be re-used, for example, in cross-validation functions, grid-searches, learning curves and so on. I will illustrate their use, and some pitfalls, in the context of a kaggle text-classification challenge.

The challenge
The goal in the StumbleUpon Evergreen …
Sql to excel
A little python tool to execute an sql script (postgresql in this case, but should be easily modifiable for mysql etc.) and store the result in a csv or excel (xls file):
"""
Executes an sql script and stores the result in a file.
"""
import os, sys
import subprocess
import csv
from xlwt import Workbook
def sql_to_csv(sql_fnm, csv_fnm):
""" Write result of executing sql script to txt file"""
with open(sql_fnm, 'r') as sql_file:
query = sql_file.read()
query = "COPY (" + query + ") TO STDOUT WITH CSV HEADER"
cmd = 'psql -c "' + query + '"'
print cmd
data = subprocess.check_output(cmd, shell=True)
with open(csv_fnm, 'wb …Retrieving your Google Scholar data
For my interactive CV I decided to try not only to automate the creation of a bibliography of my publications, but also to extend it with a citation count for each paper, which Google Scholar happens to keep track of. Unfortunately there is no Scholar API. But I figured since my own profile is based on data I essentially donated to Google, it is only fair that I can have access to it too. Hence I wrote a little scraper that iterates over the publications in my Scholar profile, extracts all citations, and bins them per year. That way I …
Tag graph plugin for Pelican
On my front page I display a sort of sitemap for my blog. Since the structure of the site is not very hierarchical, I decided to show pages and posts as a graph along with their tags. To do so, I created a mini plugin for the Pelican static blog engine. The plugin is essentially a sort of callback that gets executed when the engine has generated all posts and pages from their markdown files. I then simply take the results and write them out in a json format that d3.js understands (a list of nodes and a list …
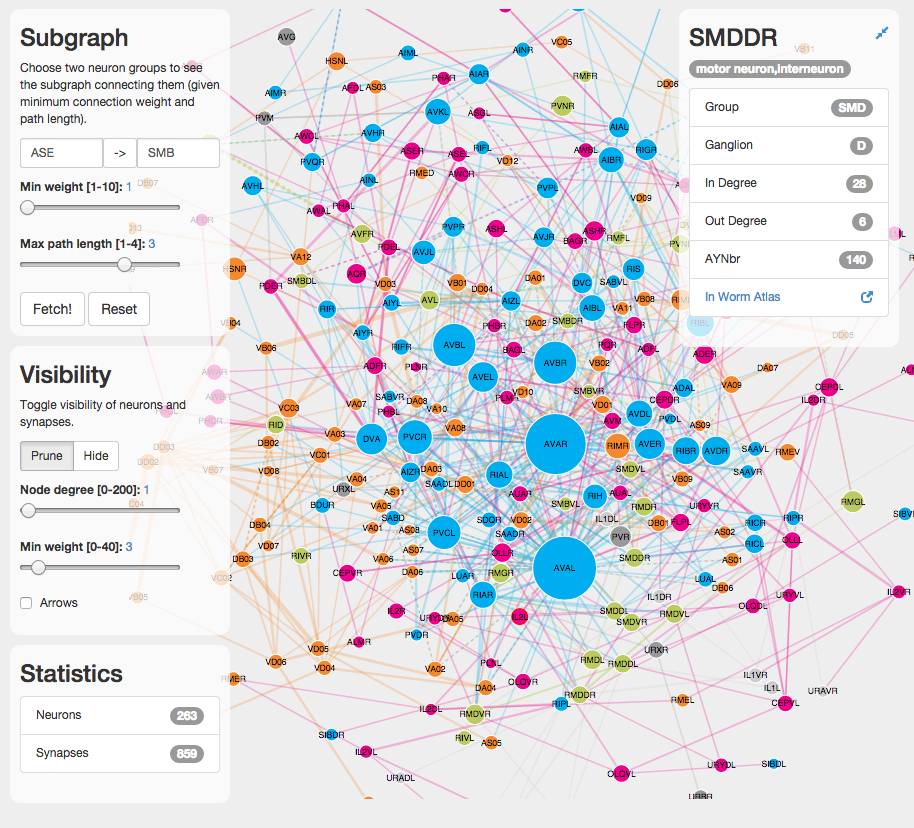
C. elegans connectome explorer
I’ve build a prototype visual exploration tool for the connectome of c. elegans. The data describing the worm’s neural network is preprocessed from publicly available information and stored as a graph database in neo4j. The d3.js visualization then fetches either the whole network or a subgraph and displays it using a force-directed layout (for now).
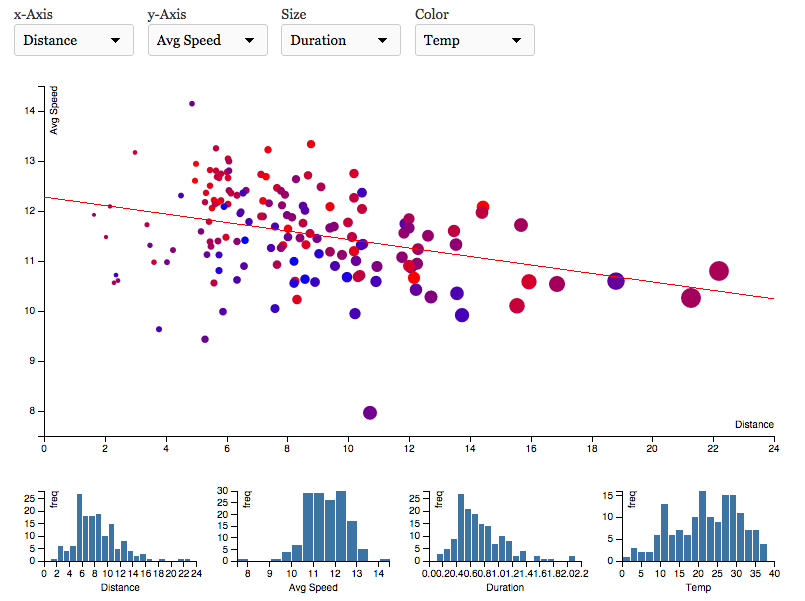
Dash+ visualization of running data
Dash+ is a python web application I built with Flask, which imports Nike+ running data into a NoSQL database (MongoDB) and uses D3.js to visualize and analyze it.
The app is work in progress and primarily intended as a personal playground for exploring d3 visualization of my own running data. Having said that, if you want to fork your own version on github, simply add your Nike access token in the corresponding file.